주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research(Neuro Technology Group)
- Center for Neuroscience Imaging Research(Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research(Neuro Technology Group)
- Center for Neuroscience Imaging Research(Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
|
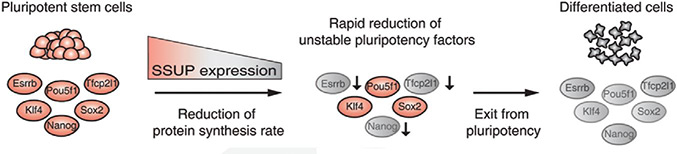
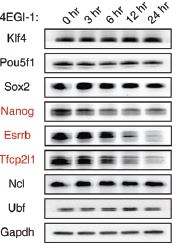
RNA’s part in determining the health of stem cells- The role of small subunit processomes in stem cell determination October 12, 2015 What makes stem cells so interesting is that they are pluripotent: they have the potential to become any cell in an organism. Their ability to differentiate into any cell and be used to replace those that are damaged or diseased holds promise, but only once the secret to controlling them has been unlocked. Working with mouse embryonic stem cells (mESCs), scientists at the IBS Center for RNA Research have come one step closer to understand how to control induced pluripotent stem cells. They found 16 RNA-binding proteins (RBPs) whose depletion results in a loss of stem cell pluripotency and uncovered six RBPs (Krr1, Ddx47, Ddx52, Nol6, Pdcd11, and Rrp7a) which make up the critical protein complex called small subunit processome (SSUP), and is responsible for making ribosomes. Their results were published this month in the journal Genes & Development. The team first set out to determine which RBPs were critical in RNA-mediated gene regulation by applying an RNAi screen to mESCs. They performed two rounds of RNAi screens against RBPs expressed in mESCs to look for pluripotency and found 16 positive hits. They discovered that the 6 RBPs that make up SSUP mediate 18S rRNA biogenesis in turn making them important regulators for pluripotent cells. Next they tested an additional 27 genes that are known to be involved in various steps of ribosome biogenesis to verify whether SSUP was required for ESC maintenance. Depletion of five SSUP proteins (Imp4, Mpp10/Mphosph10, Wdr36, Wdr46, and Wdr75) resulted in a reduction of Nanog expression, and the absence of one SSUP component (Cirh1a) caused cell death. In ESCs, SSUP subunits are up-regulated which enhances their translation rate and supports the interconnected regulatory network which controls pluripotency. The SSUP genes, including Krr1, are highly expressed in proliferating cells, including stem cells and progenitor cells, especially in ESC lines. These enhance the global translational rate, and are critical to sustain the protein levels of pluripotency factors such as Nanog and Esrrb, which can otherwise easily be disrupted. To test how translation affected ESCs, the team treated mESCs with a translational inhibitor, 4EGI-1, which resulted in Nanog, Esrrb, and Tfcp2l1 protein levels decreasing rapidly in response to translational repression. The team further treated mESCs with 4EGI-1 for 3 days at which time the mESCs lost their stem cell identity. Kwon Tae You explains “4EGI-1 treatment reduced the protein synthesis rate by about 50%,” and when the cells were re-seeded in fresh ESC medium, the 4EGI-1-treated cells showed a reduced capacity to re-establish colonies. These results show that enhanced translational activity is critical for ESC maintenance. Furthermore, the SSUP proteins are required for efficient reprogramming of induced pluripotent stem cells. When they performed the reprogramming experiment, they observed Induction of endogenous Nanog and Esrrb was substantially delayed and suppressed in Krr1-depleted cells compared to control cells.
The IBS team also uncovered that ESCs stay in a pluripotent state when the master transcription factors (TFs) are sustained at appropriate levels but when the TFs are no longer available, the cells enter into a differentiation state. This indicates that precise regulation of translation rates may critically influence the final determination of stem cells.
Exhaustive research has gone into studying the transcriptional network, while before this study post-transcriptional regulatory pathways have been relatively neglected. It is well known that ribosome biogenesis is a key component of the cell cycle as it regulates cell size and growth and the IBS team has uncovered that SSUPs play a critical role in maintaining induced pluripotent stem cell integrity. This study provides experimental evidence which shows the important role of RNA levels in the controlling the fate of embryonic stem cells, and shows an understanding of RNA's ability to differentiate stem cells at the molecular level. Additionally, since ribosomal RNA has been shown to have specified controlled of cellular fate, this study provides a theoretical basis for disease therapy and neuroscience research and may lead to future advances in treating degenerative diseases or even brain cancers. By Daniel Kopperud Notes for editors - References - Media Contact - About the Institute for Basic Science (IBS)
|
|||
|
|
| Next | |
|---|---|
| before |
- Content Manager
- Public Relations Team : Yim Ji Yeob 042-878-8173
- Last Update 2023-11-28 14:20